Main sampling algorithm of CAR-LASSO model
CARlasso( formula, data, link = "identity", adaptive = FALSE, r_beta = ifelse(adaptive, 0.01, 1), delta_beta = ifelse(adaptive, 1e-06, 0.01), r_Omega = ifelse(adaptive, 0.01, 1), delta_Omega = ifelse(adaptive, 1e-06, 0.01), lambda_diag = 0, n_iter = 2000, n_burn_in = 1000, thin_by = 10, ns = 1000, m = 20, emax = 64, progress = TRUE, verbos = TRUE )
Arguments
| formula | A double sided formula with response at left hand side and predictors at right hand side |
|---|---|
| data | A data.frame with all response and predictors, row as observations |
| link | String name of link function? Currently can be "identity" for normal response, "probit" for binary, "log" for counting, "logit" for compositional. Note that when use "logit", the last response will be used as reference. |
| adaptive | Bool, whether run the adaptive version of the model |
| r_beta | Hyper-parameter for regression coefficient, shape parameter of Gamma, if adaptive, should have row number same as number of predictors while column number of responses |
| delta_beta | Hyper-parameter for regression coefficient, rate parameter of Gamma, if adaptive, should have row number same as number of predictors while column number of responses |
| r_Omega | Hyper-parameter for precision matrix, shape parameter of Gamma. If adaptive, can be a matrix with same size as precision matrix, if this is the case, only upper triangular part without diagonal will be used, or can be a vector whose size was the upper triangular part of precision matrix, if non-adaptive, a scalar. |
| delta_Omega | Hyper-parameter for precision matrix, rate parameter of Gamma, If adaptive, can be a matrix with same size as precision matrix, if this is the case, only upper triangular part without diagonal will be used, or can be a vector whose size was the upper triangular part of precision matrix, if non-adaptive, a scalar. |
| lambda_diag | adaptive only hyper-parameter for penalties on diagonal entries of Omega, should have dimension k and non-negative |
| n_iter | Number of sampling iterations (i.e. after burn in) for the Gibbs sampler |
| n_burn_in | Number of burn in iterations for the Gibbs sampler |
| thin_by | Final sample was thin by this number |
| ns | parameter for ARS, maximum number of hulls, only used when link is "log" and "logit" |
| m | parameter for ARS, initial number of hulls, only used when link is "log" and "logit" |
| emax | parameter for ARS, tolerance for small values being 0, larger meaning we tolerate smaller values, only used when link is "log" and "logit" |
| progress | Bool, whether report progress from C++ |
| verbos | Bool, whether show warnings and messages. |
Value
A carlasso_out object with elements:
$point_est$Omega: Posterior mean of precision matrix$beta: Posterior mean of regression coefficient$CAR$C: The conditional regression coefficients among responses$B: The conditional regression coefficients between response and predictors$M: The conditional variance
$nodes$responses: node name of responses$predictors: node name of predictors
$data$response: response matrix$design: design matrix
$settings: all settings sent to the algorithm, exclude data$MCMC_output$beta: A coda::mcmc object, each row was an MCMC sample of the (column) vectorization of regression coefficient B$mu: A coda::mcmc object, each row was an MCMC sample of the mean vector$Omega: A coda::mcmc object, each row was an MCMC sample of the upper triangular part (with diagonal) of precision matrix Omega$lambda: Non-adaptive only, A coda::mcmc object, first column was the shrinkage parameter lambda for regression coefficient and the second column was shrinkage parameter lambda for precision matrix$lambda_beta: Adaptive only, A coda::mcmc object, each row was an MCMC sample of the (column) vectorization of shrinkage parameter for regression coefficient B$lambda_Omega: Adaptive only, A coda::mcmc object, each row was an MCMC sample of the shrinkage parameter for the upper triangular part (without diagonal) of precision matrix Omega
Examples
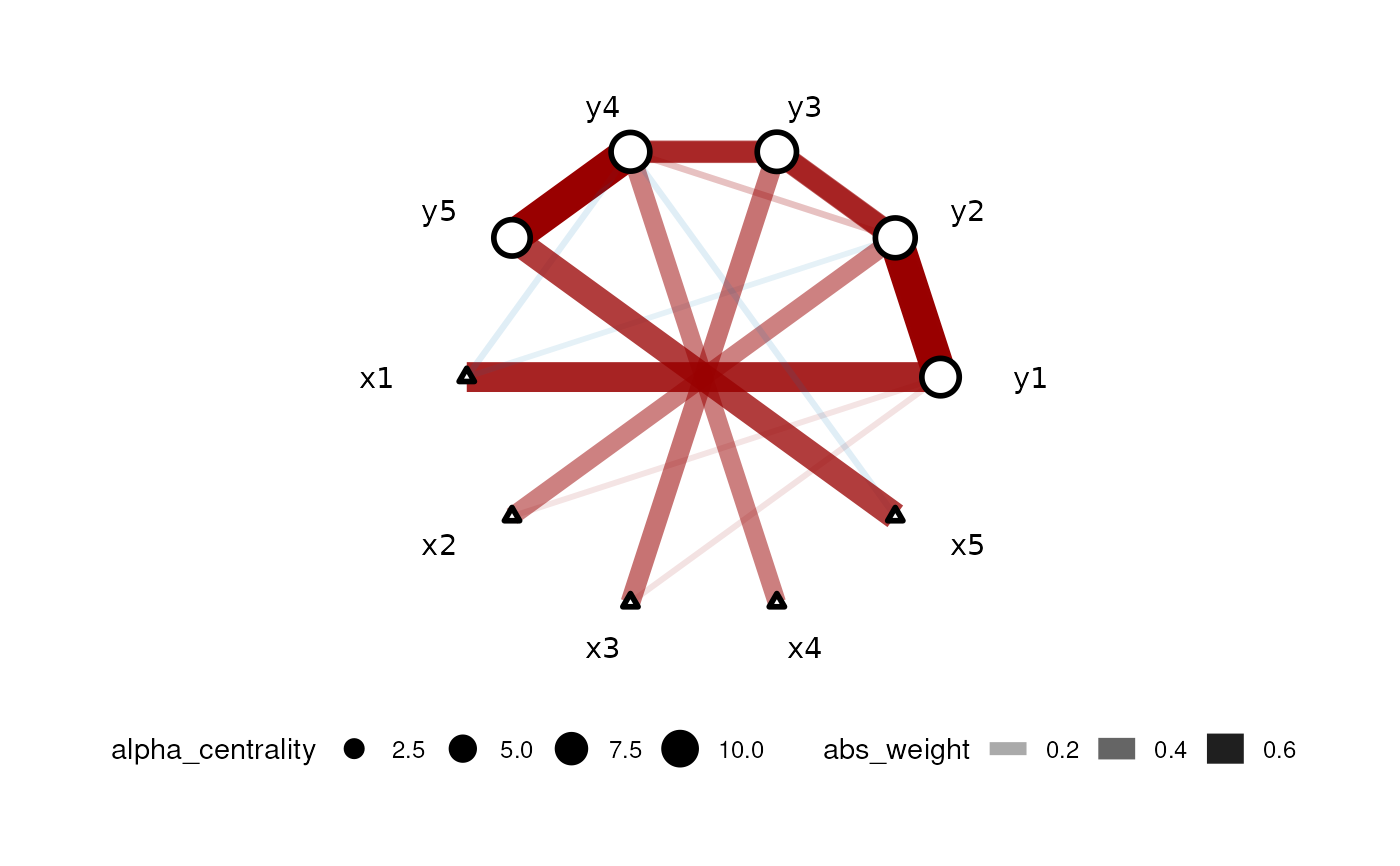
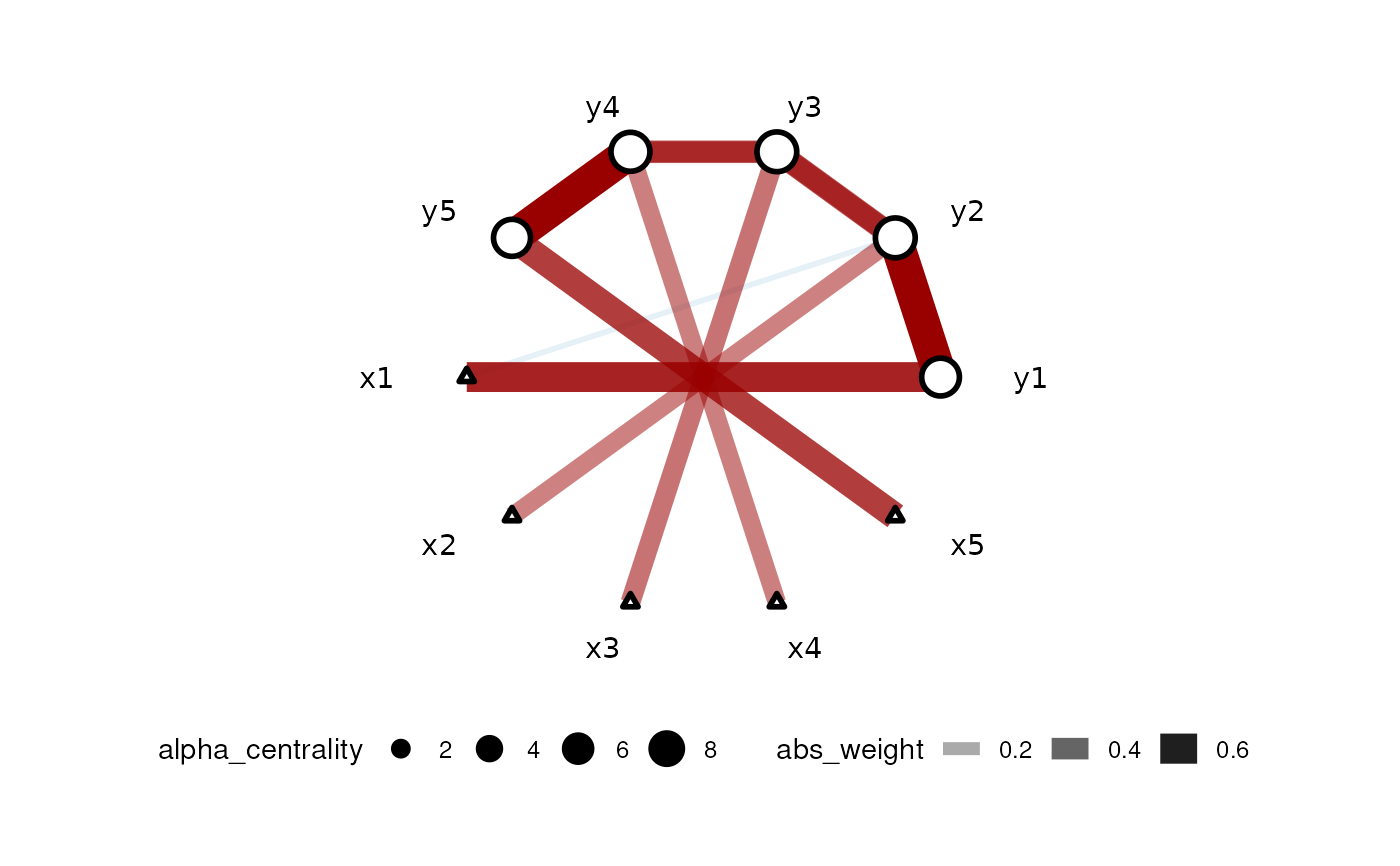
set.seed(42) dt <- simu_AR1() car_res <- CARlasso(y1+y2+y3+y4+y5~x1+x2+x3+x4+x5, data = dt, adaptive = TRUE)#> Predictors will be centered. #> #> Algorithm set to be adapive. Assuming all hyper parameters are the same for beta #> #> Algorithm set to be adapive. Assuming all hyper parameters are the same for Omega's off diagonal entries #> #> Algorithm set to be adaptive. Assuming priors are all the same for Omega's diagonals #> #> Algorithm start... #> #> progress: #> #> #> done #>