CARlasso
Published in CRAN, 2021
R implementation of
Shen, Yunyi, and Claudia Solis-Lemus. “Bayesian Conditional Auto-Regressive LASSO Models to Learn Sparse Networks with Predictors.” arXiv preprint arXiv:2012.08397 (2020).
Documentations can be found: https://yunyishen.ml/CAR-LASSO/
The package is on CRAN, to install it, use:
install.packages("CARlasso")
To install from Github:
devtools::install_github("YunyiShen/CAR-LASSO")
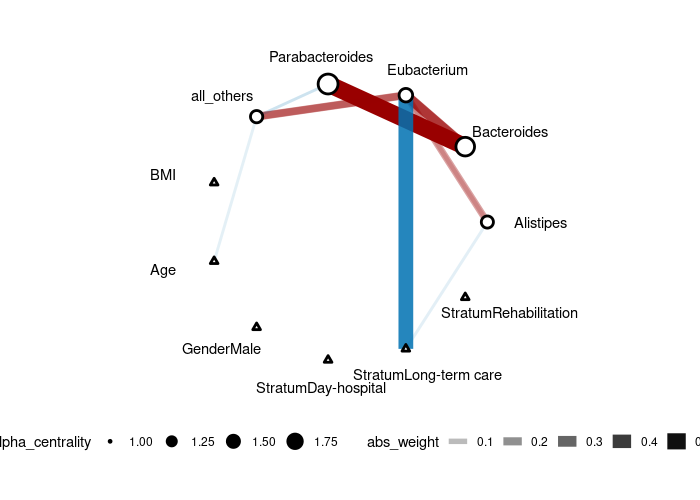
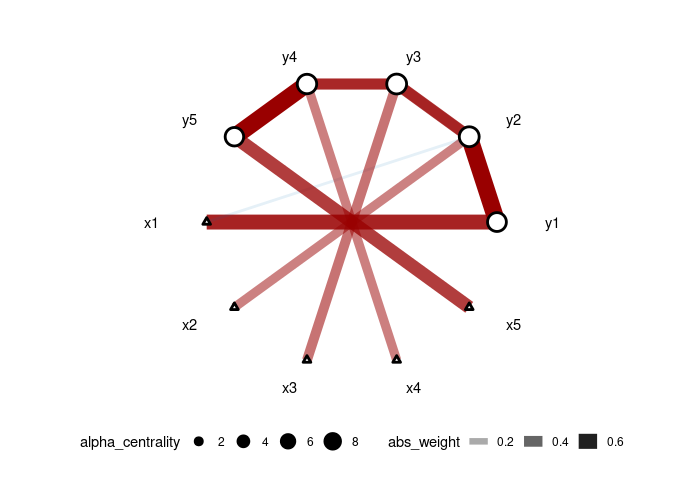
Example run, we generated data from a 5-node AR1 model with each node has a specific treatment then use CAR-ALASSO to reconstruct such network and plot the result.
set.seed(42)
dt <- simu_AR1(n=100,k=5, rho=0.7)
car_res <- CARlasso(y1+y2+y3+y4+y5~x1+x2+x3+x4+x5, data = dt, adaptive = TRUE)
plot(car_res,tol = 0.05)
# with horseshoe inference
car_res <- horseshoe(car_res)
plot(car_res)
To run a reduced version of the analysis on human gut microbiome (with less predictors and responses), try:
gut_res <- CARlasso(Alistipes+Bacteroides+
Eubacterium+Parabacteroides+all_others~
BMI+Age+Gender+Stratum,
data = mgp154,link = "logit",
adaptive = TRUE, n_iter = 5000,
n_burn_in = 1000, thin_by = 10)
# horseshoe will take a while, as it's currently implemented in R rather than C++
gut_res <- horseshoe(gut_res)
plot(gut_res)
It might take a little while due to the sampling process of the latent normal variable
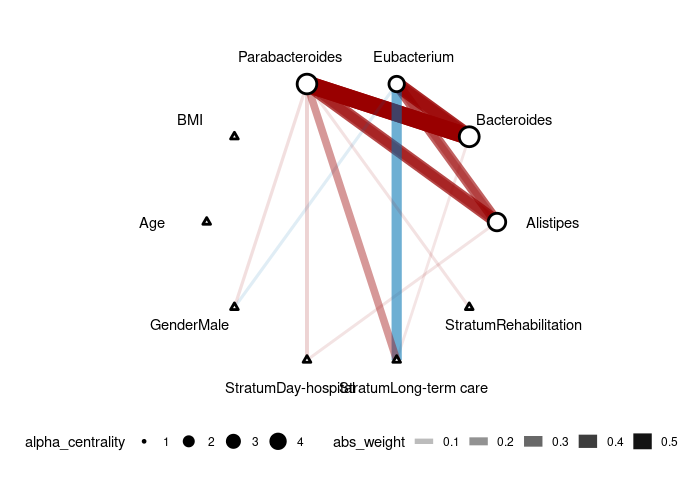
Though we don’t recommend treating compositional data as counts, as a illustration, we can run the counting model:
gut_res <- CARlasso(Alistipes+Bacteroides+
Eubacterium+Parabacteroides+all_others~
BMI+Age+Gender+Stratum,
data = mgp154,link = "log",
adaptive = TRUE,
r_beta = 0.1, # default sometimes cause singularity in Poisson model due to exponential transformation, slightly change can fix it.
n_iter = 5000,
n_burn_in = 1000, thin_by = 10)
# horseshoe will take a while, as it's currently implemented in R rather than C++
gut_res <- horseshoe(gut_res)
plot(gut_res)